The Geomicrobiology Lab, supported by CIGLR and NOAA-GLERL through the NOAA ‘Omics program, is developing the Great Lakes Atlas for Multi-omics Research (GLAMR). ‘Omics techniques allow researchers to directly characterize molecules like DNA, RNA, or metabolites in samples to determine which organisms and genes are present from DNA with metagenomics, how those genes are being expressed by studying RNA with metatranscriptomics, and what compounds are being produced with metabolomics. GLAMR is a next-generation research database that unites and standardizes ‘omics datasets and other associated environmental measurements collected by researchers across the Great Lakes region and provides a framework to study complex relationships between these datasets, including networks of interactions between organisms and ecosystems. GLAMR facilitates exploration of ‘omics datasets and enables comparison across studies using standardized bioinformatics pipelines. GLAMR supports NOAA missions such as combating harmful algal blooms and invasive species, discovering pharmaceuticals and other beneficial compounds, and protecting vulnerable species and habitats.
Understanding harmful algal blooms through ‘omics
Large harmful algal blooms (HABs) occur annually in the Laurentian Great Lakes, particularly in the western basin of Lake Erie. HABs occur when certain types of algae or cyanobacteria grow rapidly in aquatic systems, often producing toxins that can harm aquatic life, ecosystems, and even pose risks to human health. ‘Omics data has become increasingly valuable in understanding and addressing issues related to these blooms, including factors influencing the production of toxins like microcystin, a hepatotoxin produced by the cyanobacteria Microcystis. Elevated levels of microcystin in the city of Toledo’s drinking water, sourced from Lake Erie, led to a do-not-drink order in 2014.
During the summer bloom season, in addition to measurements of water quality and nutrients like nitrogen and phosphorus, researchers collect samples by filtering lake water through filters to concentrate cells, from which DNA or RNA is extracted and sequenced. Various computational tools are then used to process these sequences and compare them to known references. For instance, the community composition of a sample can be determined by evaluating the percent of sequences corresponding to each organism, while correcting for differences in genome size. Further, the genes involved in biosynthesis of several key toxins are known, and by evaluating the number of sequences corresponding to each, their abundance—and thus the potential for toxin production–can be determined. Notably, the presence of toxigenic genes is only loosely correlated with actual toxin production, as organisms capable of producing a given toxin are not always doing so; controls on toxin production are an active area of research. Researchers also use this information to better understand differences in the species and strains responsible for blooms, potential impacts of climate change, and to evaluate emerging threats such as novel toxins.
‘Omics on Seagull
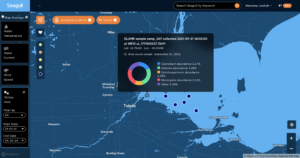
Recognizing the importance of accessibility, the team has prioritized user-friendly access through a public interface, catering to both seasoned experts and those less familiar with bioinformatics or ‘omics methodologies. To achieve this, a subset of key HABs-specific ‘omics data from GLAMR is now conveniently accessible via a new Seagull ‘omics layer, a simple-to-use map designed for seamless integration of diverse information. The ‘omics layer provides access to abundance information for organism and toxin-biosynthesis genes, geographical information, and environmental and water quality information. The overarching objective is to democratize access to the database, enabling a broad spectrum of users to explore and extract valuable insights. Through Seagull’s map-based interface, users can effortlessly initiate queries, view graphical data summaries, and thus streamline the process of ‘omics research.
The initial phase of this endeavor focuses on the development of the ‘Omics Basic Discovery app, tailored to enhance forecasts of Harmful Algal Blooms (HABs) in the Great Lakes region, thereby safeguarding both the environmental health and economic prosperity of the area. This app empowers users to delve into genetic data pertaining to a subset of organisms linked to HABs, spatially and temporally across specific HABs sites, all through a map-based layer on Seagull. Users can visualize the relative abundances of HAB-related organisms at sampling sites from 2010 onwards through interactive stacked bar plots and also simultaneously visualize environmental and water quality data from the samples.
For those keen on deeper exploration, the full datasets are readily accessible on the GLAMR website, ensuring comprehensive access to valuable ‘Omics data for all interested parties.
Information and content contributions provided by: Sneha Bhadbhade and Anders Kiledal